Overview
The ImmPort Data Model represents evolutionary work within the immunology research community by supporting multiple data standards, ontologies, and interfaces to promote FAIR data sharing. Details develop the data model is captured in the 2012 publication Toward an Ontology-Based Framework for Clinical Research Databases. The Basic Formal Ontology (BFO) and Ontology for Biomedical Investigations (OBI) were used as the foundation for the classes of entities and relationships represented in the model. The model also incorporates clinical data standards from organizations such as the Clinical Data Interchange Standards Consortium (CDISC).
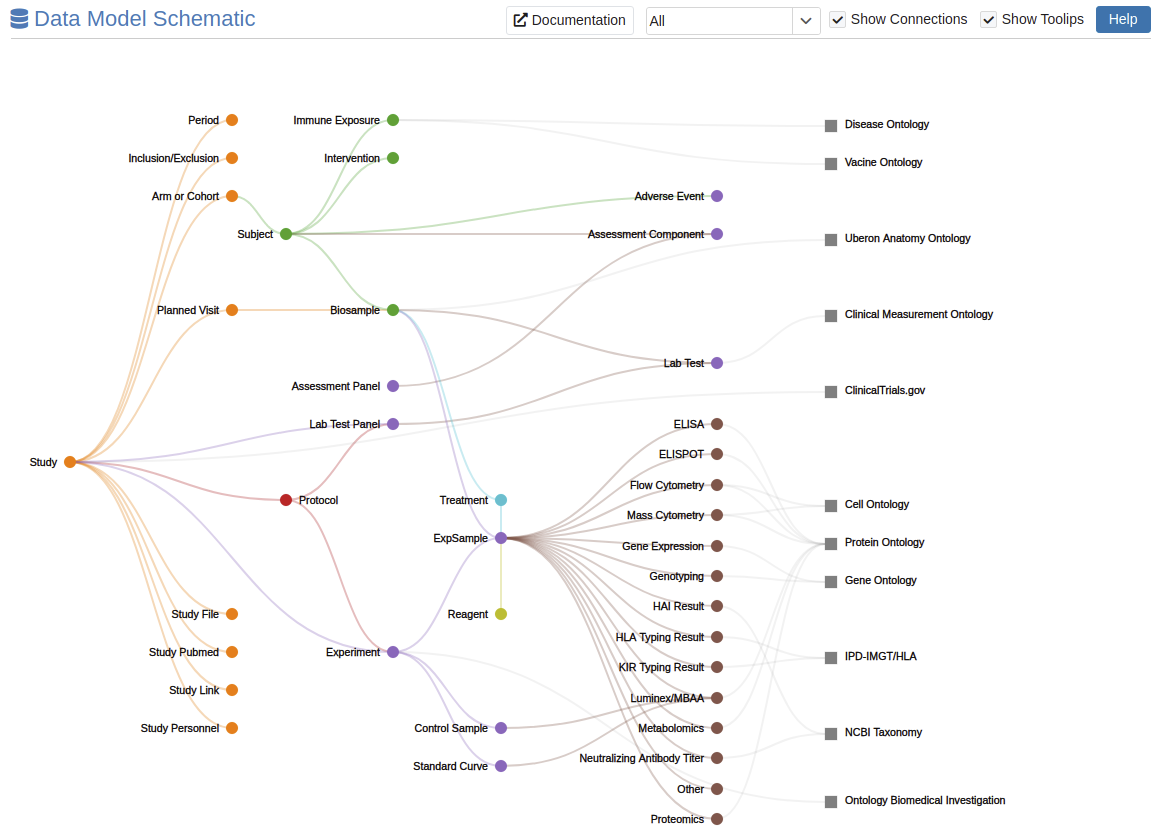
As seen in the below graphic, the logical model format is stored as a Relational Database Model, where tables capture information for a specific feature, such as a subject or biosample, and related tables are linked through foreign keys. Data is first organized primarily into a study table; the study and its attendant protocols are foundational elements of the scientific method to explain what will be studied, and with what approaches. Each study can be linked to multiple human or animal subject participants in a study, as well as relevant results data on each subject. Data collected about study subjects may take many forms and formats, including questionnaire-based data, clinical trial outcomes, lab tests, medications, and/or mechanistic assays.
An interactive graphic of the ImmPort Data Model is available. Click on image to view.